lamindb.Artifact¶
- class lamindb.Artifact(data: UPathStr, kind: ArtifactKind | None = None, key: str | None = None, description: str | None = None, revises: Artifact | None = None, run: Run | None = None)¶
Bases:
DBRecord,IsVersioned,TracksRun,TracksUpdatesDatasets & models stored as files, folders, or arrays.
Artifacts manage data in local or remote storage.
Some artifacts are array-like, e.g., when stored as
.parquet,.h5ad,.zarr, or.tiledb.- Parameters:
data –
UPathStrA path to a local or remote folder or file.kind –
Literal["dataset", "model"] | None = NoneDistinguish models from datasets from other files & folders.key –
str | None = NoneA path-like key to reference artifact in default storage, e.g.,"myfolder/myfile.fcs". Artifacts with the same key form a version family.description –
str | None = NoneA description.revises –
Artifact | None = NonePrevious version of the artifact. Is an alternative way to passingkeyto trigger a new version.run –
Run | None = NoneThe run that creates the artifact.
Examples
Create an artifact from a local file or folder:
artifact = ln.Artifact("./my_file.parquet", key="examples/my_file.parquet").save() artifact = ln.Artifact("./my_folder", key="project1/my_folder").save()
Calling
.save()copies or uploads the file to the default storage location of your lamindb instance. If you create an artifact from a remote file or folder, lamindb merely registers the S3keyand avoids copying the data:artifact = ln.Artifact("s3://my_bucket/my_folder/my_file.csv").save()
If you want to validate & annotate an array, pass a
schemato one of the.from_df(),.from_anndata(), … constructors:schema = ln.Schema(itype=ln.Feature) # a schema that merely enforces that feature names exist in the Feature registry artifact = ln.Artifact.from_df("./my_file.parquet", key="my_dataset.parquet", schema=schema).save() # validated and annotated
You can make a new version of an artifact by passing an existing
key:artifact_v2 = ln.Artifact("./my_file.parquet", key="examples/my_file.parquet").save() artifact_v2.versions.df() # see all versions
You can write artifacts to other storage locations by switching the current default storage location (
storage):ln.settings.storage = "s3://some-bucket"
Sometimes you want to avoid mapping the artifact into a path hierarchy, and you only pass
description:artifact = ln.Artifact("./my_folder", description="My folder").save() artifact_v2 = ln.Artifact("./my_folder", revises=old_artifact).save() # need to version based on `revises`, a shared description does not trigger a new version
Notes
Typical storage formats & their API accessors
Arrays:
Table:
.csv,.tsv,.parquet,.ipc⟷DataFrame,pyarrow.TableAnnotated matrix:
.h5ad,.h5mu,.zrad⟷AnnData,MuDataGeneric array: HDF5 group, zarr group, TileDB store ⟷ HDF5, zarr, TileDB loaders
Non-arrays:
Image:
.jpg,.png⟷np.ndarray, …Fastq:
.fastq⟷ /VCF:
.vcf⟷ /QC:
.html⟷ /
You’ll find these values in the
suffix&accessorfields.LaminDB makes some default choices (e.g., serialize a
DataFrameas a.parquetfile).Will artifacts get duplicated?
If an artifact with the exact same hash already exists,
Artifact()returns the existing artifact.In concurrent workloads where the same artifact is created repeatedly at the exact same time,
.save()detects the duplication and will return the existing artifact.Why does the constructor look the way it looks?
It’s inspired by APIs building on AWS S3.
Both boto3 and quilt select a bucket (a storage location in LaminDB) and define a target path through a
keyargument.In boto3:
# signature: S3.Bucket.upload_file(filepath, key) import boto3 s3 = boto3.resource('s3') bucket = s3.Bucket('mybucket') bucket.upload_file('/tmp/hello.txt', 'hello.txt')
In quilt3:
# signature: quilt3.Bucket.put_file(key, filepath) import quilt3 bucket = quilt3.Bucket('mybucket') bucket.put_file('hello.txt', '/tmp/hello.txt')
See also
StorageStorage locations for artifacts.
CollectionCollections of artifacts.
from_df()Create an artifact from a
DataFrame.from_anndata()Create an artifact from an
AnnData.
Attributes¶
- DoesNotExist = <class 'lamindb.models.artifact.Artifact.DoesNotExist'>¶
- Meta = <class 'lamindb.models.dbrecord.DBRecord.Meta'>¶
- MultipleObjectsReturned = <class 'lamindb.models.artifact.Artifact.MultipleObjectsReturned'>¶
- biologics¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- biosamples¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- cell_lines¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- cell_markers¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- cell_types¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- clinical_trials¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- clinicore_biosamples¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- collections: Collection¶
The collections that this artifact is part of.
- combination_perturbations¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- compound_perturbations¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- compounds¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- created_by: User¶
Creator of record.
- created_by_id¶
- developmental_stages¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- diseases¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- donors¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- environmental_perturbations¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- ethnicities¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- experimental_factors¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- experiments¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- feature_sets: Schema¶
The feature sets measured by the artifact.
- features(host): FeatureManager = <class 'lamindb.models._feature_manager.FeatureManagerArtifact'>¶
Feature manager.
Typically, you annotate a dataset with features by defining a
Schemaand passing it to theArtifactconstructor.Here is how to do annotate an artifact ad hoc:
artifact.features.add_values({ "species": organism, # here, organism is an Organism record "scientist": ['Barbara McClintock', 'Edgar Anderson'], "temperature": 27.6, "experiment": "Experiment 1" })
Query artifacts by features:
ln.Artifact.filter(scientist="Barbara McClintock")
Features may or may not be part of the artifact content in storage. For instance, the
DataFrameCuratorflow validates the columns of aDataFrame-like artifact and annotates it with features corresponding to these columns.artifact.features.add_values, by contrast, does not validate the content of the artifact. Attributes ———-Class methods¶
- classmethod filter(*queries, **expressions)¶
Query a set of artifacts.
- Parameters:
*queries –
Qexpressions.**expressions – Features & fields via the Django query syntax.
- Return type:
See also
Guide: Query & search registries
Examples
Query by fields:
ln.Arfifact.filter(key="examples/my_file.parquet")
Query by features:
ln.Arfifact.filter(cell_type_by_model__name="T cell")
- classmethod get(idlike=None, *, is_run_input=False, **expressions)¶
Get a single artifact.
- Parameters:
idlike (
int|str|None, default:None) – Either a uid stub, uid or an integer id.is_run_input (
bool|Run, default:False) – Whether to track this artifact as run input.expressions – Fields and values passed as Django query expressions.
- Raises:
lamindb.errors.DoesNotExist – In case no matching record is found.
- Return type:
See also
Guide: Query & search registries
Method in
DBRecordbase class:get()
Examples
artifact = ln.Artifact.get("tCUkRcaEjTjhtozp0000") artifact = ln.Arfifact.get(key="examples/my_file.parquet")
Methods¶
- describe(return_str=False)¶
Describe record including linked records.
- Parameters:
return_str (
bool, default:False) – Return a string instead of printing.- Return type:
None
- genes¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- genetic_perturbations¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- input_of_runs: Run¶
Runs that use this artifact as an input.
- property labels: LabelManager¶
Label manager.
To annotate with labels, you typically use the registry-specific accessors, for instance
ulabels:experiment = ln.ULabel(name="Experiment 1").save() artifact.ulabels.add(experiment)
Similarly, you query based on these accessors:
ln.Artifact.filter(ulabels__name="Experiment 1").all()
Unlike the registry-specific accessors, the
.labelsaccessor provides a way of associating labels with features:experiment = ln.Feature(name="experiment", dtype="cat").save() artifact.labels.add(experiment, feature=study)
Note that the above is equivalent to:
artifact.features.add_values({"experiment": experiment})
- links_biologic¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_biosample¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_cell_line¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_cell_marker¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_cell_type¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_clinical_trial¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_collection¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_combination_perturbation¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_compound¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_compound_perturbation¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_developmental_stage¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_disease¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_donor¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_environmental_perturbation¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_ethnicity¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_experiment¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_experimental_factor¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_featurevalue¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_gene¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_genetic_perturbation¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_medication¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_organism¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_pathway¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_patient¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_perturbation_target¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_phenotype¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_project¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_protein¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_reference¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_techsample¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_tissue¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_treatment¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_ulabel¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- links_well¶
Accessor to the related objects manager on the reverse side of a many-to-one relation.
In the example:
class Child(Model): parent = ForeignKey(Parent, related_name='children')
Parent.childrenis aReverseManyToOneDescriptorinstance.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- medications¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- objects = <lamindb.models.query_manager.QueryManager object>¶
- organisms¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- property path: Path¶
Path.
Example:
import lamindb as ln # File in cloud storage, here AWS S3: artifact = ln.Artifact("s3://my-bucket/my-file.csv").save() artifact.path #S3QueryPath('s3://my-bucket/my-file.csv') # File in local storage: ln.Artifact("./myfile.csv", key="myfile.csv").save() artifact.path #> PosixPath('/home/runner/work/lamindb/lamindb/docs/guide/mydata/myfile.csv')
- pathways¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- patients¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- perturbation_targets¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- phenotypes¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- property pk¶
- projects: Project¶
Linked projects.
- proteins¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- references: Reference¶
Linked references.
- run: Run | None¶
Run that created the artifact.
- run_id¶
- schema_id¶
- space: Space¶
The space in which the record lives.
- space_id¶
- property stem_uid: str¶
Universal id characterizing the version family.
The full uid of a record is obtained via concatenating the stem uid and version information:
stem_uid = random_base62(n_char) # a random base62 sequence of length 12 (transform) or 16 (artifact, collection) version_uid = "0000" # an auto-incrementing 4-digit base62 number uid = f"{stem_uid}{version_uid}" # concatenate the stem_uid & version_uid
- storage: Storage¶
Storage location, e.g. an S3 or GCP bucket or a local directory.
- storage_id¶
- techsamples¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- tissues¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- treatments¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
- property versions: QuerySet¶
Lists all records of the same version family.
>>> new_artifact = ln.Artifact(df2, revises=artifact).save() >>> new_artifact.versions()
- wells¶
Accessor to the related objects manager on the forward and reverse sides of a many-to-many relation.
In the example:
class Pizza(Model): toppings = ManyToManyField(Topping, related_name='pizzas')
Pizza.toppingsandTopping.pizzasareManyToManyDescriptorinstances.Most of the implementation is delegated to a dynamically defined manager class built by
create_forward_many_to_many_manager()defined below.
Class methods¶
- classmethod filter(*queries, **expressions)¶
Query a set of artifacts.
- Parameters:
*queries –
Qexpressions.**expressions – Features & fields via the Django query syntax.
- Return type:
See also
Guide: Query & search registries
Examples
Query by fields:
ln.Arfifact.filter(key="examples/my_file.parquet")
Query by features:
ln.Arfifact.filter(cell_type_by_model__name="T cell")
- classmethod from_anndata(adata, *, key=None, description=None, run=None, revises=None, schema=None, **kwargs)¶
Create from
AnnData, optionally validate & annotate.- Parameters:
adata (
AnnData| lamindb.core.types.UPathStr) – AnAnnDataobject or a path of AnnData-like.key (
str|None, default:None) – A relative path within default storage, e.g.,"myfolder/myfile.h5ad".description (
str|None, default:None) – A description.revises (
Artifact|None, default:None) – An old version of the artifact.run (
Run|None, default:None) – The run that creates the artifact.schema (
Schema|None, default:None) – A schema that defines how to validate & annotate.
- Return type:
See also
Collection()Track collections.
FeatureTrack features.
Example
No validation and annotation:
import lamindb as ln adata = ln.core.datasets.anndata_with_obs() artifact = ln.Artifact.from_anndata(adata, key="mini_anndata_with_obs.h5ad").save()
With validation and annotation.
import lamindb as ln ln.core.datasets.mini_immuno.define_features_labels() adata = ln.core.datasets.mini_immuno.get_dataset1(otype="AnnData") schema = ln.examples.schemas.anndata_ensembl_gene_ids_and_valid_features_in_obs() artifact = ln.Artifact.from_anndata( adata, key="examples/mini_immuno.h5ad", schema=schema ).save() artifact.describe()
Under-the-hood, this used the following schema.
import lamindb as ln import bionty as bt obs_schema = ln.examples.schemas.valid_features() varT_schema = ln.Schema( name="valid_ensembl_gene_ids", itype=bt.Gene.ensembl_gene_id ).save() schema = ln.Schema( name="anndata_ensembl_gene_ids_and_valid_features_in_obs", otype="AnnData", slots={"obs": obs_schema, "var.T": varT_schema}, ).save()
This schema tranposes the
varDataFrame during curation, so that one validates and annotates thevar.Tschema, i.e.,[ENSG00000153563, ENSG00000010610, ENSG00000170458]. If one doesn’t transpose, one would annotate with the schema ofvar, i.e.,[gene_symbol, gene_type].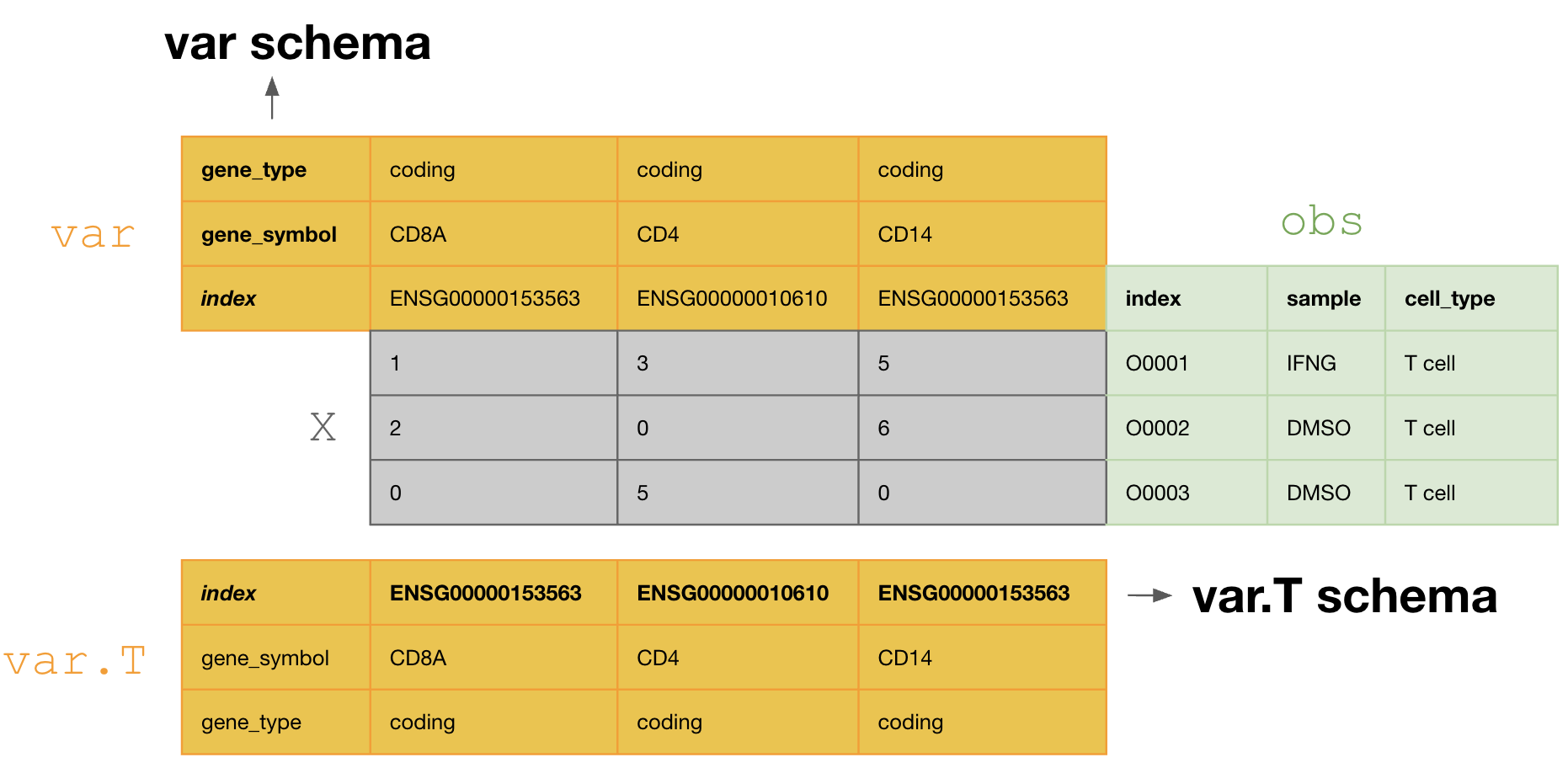
- classmethod from_df(df, *, key=None, description=None, run=None, revises=None, schema=None, **kwargs)¶
Create from
DataFrame, optionally validate & annotate.- Parameters:
df (
DataFrame) – ADataFrameobject.key (
str|None, default:None) – A relative path within default storage, e.g.,"myfolder/myfile.parquet".description (
str|None, default:None) – A description.revises (
Artifact|None, default:None) – An old version of the artifact.run (
Run|None, default:None) – The run that creates the artifact.schema (
Schema|None, default:None) – A schema that defines how to validate & annotate.
- Return type:
See also
Collection()Track collections.
FeatureTrack features.
Example
No validation and annotation:
import lamindb as ln df = ln.core.datasets.mini_immuno.get_dataset1() artifact = ln.Artifact.from_df(df, key="examples/dataset1.parquet").save()
With validation and annotation.
import lamindb as ln ln.core.datasets.mini_immuno.define_features_labels() schema = ln.examples.schemas.valid_features() df = ln.core.datasets.small_dataset1(otype="DataFrame") artifact = ln.Artifact.from_df( df, key="examples/dataset1.parquet", schema=schema ).save() artifact.describe()
Under-the-hood, this used the following schema.
import lamindb as ln schema = ln.Schema(name="valid_features", itype=ln.Feature).save()
Valid features & labels were defined as:
import lamindb as ln import bionty as bt # define valid labels perturbation_type = ln.ULabel(name="Perturbation", is_type=True).save() ln.ULabel(name="DMSO", type=perturbation_type).save() ln.ULabel(name="IFNG", type=perturbation_type).save() bt.CellType.from_source(name="B cell").save() bt.CellType.from_source(name="T cell").save() # define valid features ln.Feature(name="perturbation", dtype=perturbation_type).save() ln.Feature(name="cell_type_by_model", dtype=bt.CellType).save() ln.Feature(name="cell_type_by_expert", dtype=bt.CellType).save() ln.Feature(name="assay_oid", dtype=bt.ExperimentalFactor.ontology_id).save() ln.Feature(name="donor", dtype=str, nullable=True).save() ln.Feature(name="concentration", dtype=str).save() ln.Feature(name="treatment_time_h", dtype="num", coerce_dtype=True).save()
- classmethod from_dir(path, *, key=None, run=None)¶
Create a list of artifact objects from a directory.
Hint
If you have a high number of files (several 100k) and don’t want to track them individually, create a single
ArtifactviaArtifact(path)for them. See, e.g., RxRx: cell imaging.- Parameters:
path (lamindb.core.types.UPathStr) – Source path of folder.
key (
str|None, default:None) – Key for storage destination. IfNoneand directory is in a registered location, the inferredkeywill reflect the relative position. IfNoneand directory is outside of a registered storage location, the inferred key defaults topath.name.run (
Run|None, default:None) – ARunobject.
- Return type:
list[Artifact]
Example:
import lamindb as ln dir_path = ln.core.datasets.generate_cell_ranger_files("sample_001", ln.settings.storage) artifacts = ln.Artifact.from_dir(dir_path) ln.save(artifacts)
- classmethod from_mudata(mdata, *, key=None, description=None, run=None, revises=None, schema=None, **kwargs)¶
Create from
MuData, optionally validate & annotate.- Parameters:
mdata (
MuData| lamindb.core.types.UPathStr) – AMuDataobject.key (
str|None, default:None) – A relative path within default storage, e.g.,"myfolder/myfile.h5mu".description (
str|None, default:None) – A description.revises (
Artifact|None, default:None) – An old version of the artifact.run (
Run|None, default:None) – The run that creates the artifact.schema (
Schema|None, default:None) – A schema that defines how to validate & annotate.
- Return type:
See also
Collection()Track collections.
FeatureTrack features.
Example:
import lamindb as ln mdata = ln.core.datasets.mudata_papalexi21_subset() artifact = ln.Artifact.from_mudata(mdata, key="mudata_papalexi21_subset.h5mu").save()
- classmethod from_spatialdata(sdata, *, key=None, description=None, run=None, revises=None, schema=None, **kwargs)¶
Create from
SpatialData, optionally validate & annotate.- Parameters:
sdata (
SpatialData| lamindb.core.types.UPathStr) – ASpatialDataobject.key (
str|None, default:None) – A relative path within default storage, e.g.,"myfolder/myfile.zarr".description (
str|None, default:None) – A description.revises (
Artifact|None, default:None) – An old version of the artifact.run (
Run|None, default:None) – The run that creates the artifact.schema (
Schema|None, default:None) – A schema that defines how to validate & annotate.
- Return type:
See also
Collection()Track collections.
FeatureTrack features.
Example
No validation and annotation:
import lamindb as ln artifact = ln.Artifact.from_spatialdata(sdata, key="my_dataset.zarr").save()
With validation and annotation.
import lamindb as ln import bionty as bt attrs_schema = ln.Schema( features=[ ln.Feature(name="bio", dtype=dict).save(), ln.Feature(name="tech", dtype=dict).save(), ], ).save() sample_schema = ln.Schema( features=[ ln.Feature(name="disease", dtype=bt.Disease, coerce_dtype=True).save(), ln.Feature( name="developmental_stage", dtype=bt.DevelopmentalStage, coerce_dtype=True, ).save(), ], ).save() tech_schema = ln.Schema( features=[ ln.Feature(name="assay", dtype=bt.ExperimentalFactor, coerce_dtype=True).save(), ], ).save() obs_schema = ln.Schema( features=[ ln.Feature(name="sample_region", dtype="str").save(), ], ).save() # Schema enforces only registered Ensembl Gene IDs are valid (maximal_set=True) varT_schema = ln.Schema(itype=bt.Gene.ensembl_gene_id, maximal_set=True).save() sdata_schema = ln.Schema( name="spatialdata_blobs_schema", otype="SpatialData", slots={ "attrs:bio": sample_schema, "attrs:tech": tech_schema, "attrs": attrs_schema, "tables:table:obs": obs_schema, "tables:table:var.T": varT_schema, }, ).save()
import lamindb as ln spatialdata = ln.core.datasets.spatialdata_blobs() sdata_schema = ln.Schema.get(name="spatialdata_blobs_schema") curator = ln.curators.SpatialDataCurator(spatialdata, sdata_schema) try: curator.validate() except ln.errors.ValidationError: pass spatialdata.tables["table"].var.drop(index="ENSG00000999999", inplace=True) # validate again (must pass now) and save artifact artifact = ln.Artifact.from_spatialdata( spatialdata, key="examples/spatialdata1.zarr", schema=sdata_schema ).save() artifact.describe()
- classmethod from_tiledbsoma(path, *, key=None, description=None, run=None, revises=None, **kwargs)¶
Create from a tiledbsoma store.
- Parameters:
path (lamindb.core.types.UPathStr) – A tiledbsoma store with .tiledbsoma suffix.
key (
str|None, default:None) – A relative path within default storage, e.g.,"myfolder/mystore.tiledbsoma".description (
str|None, default:None) – A description.revises (
Artifact|None, default:None) – An old version of the artifact.run (
Run|None, default:None) – The run that creates the artifact.
- Return type:
Example:
import lamindb as ln artifact = ln.Artifact.from_tiledbsoma("s3://mybucket/store.tiledbsoma", description="a tiledbsoma store").save()
- classmethod get(idlike=None, *, is_run_input=False, **expressions)¶
Get a single artifact.
- Parameters:
idlike (
int|str|None, default:None) – Either a uid stub, uid or an integer id.is_run_input (
bool|Run, default:False) – Whether to track this artifact as run input.expressions – Fields and values passed as Django query expressions.
- Raises:
lamindb.errors.DoesNotExist – In case no matching record is found.
- Return type:
See also
Guide: Query & search registries
Method in
DBRecordbase class:get()
Examples
artifact = ln.Artifact.get("tCUkRcaEjTjhtozp0000") artifact = ln.Arfifact.get(key="examples/my_file.parquet")
Methods¶
- async adelete(using=None, keep_parents=False)¶
- async arefresh_from_db(using=None, fields=None, from_queryset=None)¶
- async asave(*args, force_insert=False, force_update=False, using=None, update_fields=None)¶
- cache(*, is_run_input=None, mute=False, **kwargs)¶
Download cloud artifact to local cache.
Follows synching logic: only caches an artifact if it’s outdated in the local cache.
Returns a path to a locally cached on-disk object (say a
.jpgfile).- Parameters:
mute (
bool, default:False) – Silence logging of caching progress.is_run_input (
bool|None, default:None) – Whether to track this artifact as run input.
- Return type:
Path
Example:
# Sync file from cloud and return the local path of the cache artifact.cache() #> PosixPath('/home/runner/work/Caches/lamindb/lamindb-ci/lndb-storage/pbmc68k.h5ad')
- clean()¶
Hook for doing any extra model-wide validation after clean() has been called on every field by self.clean_fields. Any ValidationError raised by this method will not be associated with a particular field; it will have a special-case association with the field defined by NON_FIELD_ERRORS.
- clean_fields(exclude=None)¶
Clean all fields and raise a ValidationError containing a dict of all validation errors if any occur.
- date_error_message(lookup_type, field_name, unique_for)¶
- delete(permanent=None, storage=None, using_key=None)¶
Trash or permanently delete.
A first call to
.delete()puts an artifact into the trash (sets_branch_codeto-1). A second call permanently deletes the artifact. If it is a folder artifact with multiple versions, deleting a non-latest version will not delete the underlying storage by default (ifstorage=Trueis not specified). Deleting the latest version will delete all the versions for folder artifacts.FAQ: Storage FAQ
- Parameters:
permanent (
bool|None, default:None) – Permanently delete the artifact (skip trash).storage (
bool|None, default:None) – Indicate whether you want to delete the artifact in storage.
- Return type:
None
Example:
import lamindb as ln # For an `Artifact` object `artifact`, call: artifact = ln.Artifact.get(key="some.csv") artifact.delete() # delete a single file artifact artifact = ln.Artifact.filter(key="some.tiledbsoma". is_latest=False).first() artiact.delete() # delete an old version, the data will not be deleted artifact = ln.Artifact.get(key="some.tiledbsoma". is_latest=True) artiact.delete() # delete all versions, the data will be deleted or prompted for deletion.
- describe(return_str=False)¶
Describe record including linked records.
- Parameters:
return_str (
bool, default:False) – Return a string instead of printing.- Return type:
None
- get_constraints()¶
- get_deferred_fields()¶
Return a set containing names of deferred fields for this instance.
- load(*, is_run_input=None, mute=False, **kwargs)¶
Cache and load into memory.
See all
loaders.- Parameters:
is_run_input (
bool|None, default:None) – Whether to track this artifact as run input.mute (
bool, default:False) – Silence logging of caching progress.**kwargs – Keyword arguments for the loader.
- Return type:
Any
Examples
Load a
DataFrame-like artifact:>>> artifact.load().head() sepal_length sepal_width petal_length petal_width iris_organism_code 0 0.051 0.035 0.014 0.002 0 1 0.049 0.030 0.014 0.002 0 2 0.047 0.032 0.013 0.002 0 3 0.046 0.031 0.015 0.002 0 4 0.050 0.036 0.014 0.002 0
Load an
AnnData-like artifact:>>> artifact.load() AnnData object with n_obs × n_vars = 70 × 765
Fall back to
cache()if no in-memory representation is configured:>>> artifact.load() PosixPath('/home/runner/work/lamindb/lamindb/docs/guide/mydata/.lamindb/jb7BY5UJoQVGMUOKiLcn.jpg')
- open(mode='r', engine='pyarrow', is_run_input=None, **kwargs)¶
Open a dataset for streaming.
Works for
AnnData(.h5adand.zarr), generichdf5andzarr,tiledbsomaobjects (.tiledbsoma),pyarroworpolarscompatible formats (.parquet,.csv,.ipcetc. files or directories with such files).- Parameters:
mode (str, default:
'r') – can only be"w"(write mode) fortiledbsomastores, otherwise should be always"r"(read-only mode).engine (Literal[‘pyarrow’, ‘polars’], default:
'pyarrow') – Which module to use for lazy loading of a dataframe frompyarroworpolarscompatible formats. This has no effect if the artifact is not a dataframe, i.e. if it is anAnnData,hdf5,zarrortiledbsomaobject.is_run_input (bool | None, default:
None) – Whether to track this artifact as run input.**kwargs – Keyword arguments for the accessor, i.e.
h5pyorzarrconnection,pyarrow.dataset.dataset,polars.scan_*function.
- Return type:
AnnDataAccessor | BackedAccessor | SOMACollection | SOMAExperiment | SOMAMeasurement | PyArrowDataset | Iterator[PolarsLazyFrame]
Notes
For more info, see guide: Slice arrays.
Example:
import lamindb as ln # Read AnnData in backed mode from cloud artifact = ln.Artifact.get(key="lndb-storage/pbmc68k.h5ad") artifact.open() #> AnnDataAccessor object with n_obs × n_vars = 70 × 765 #> constructed for the AnnData object pbmc68k.h5ad #> ... artifact = ln.Artifact.get(key="lndb-storage/df.parquet") artifact.open() #> pyarrow._dataset.FileSystemDataset
- prepare_database_save(field)¶
- refresh_from_db(using=None, fields=None, from_queryset=None)¶
Reload field values from the database.
By default, the reloading happens from the database this instance was loaded from, or by the read router if this instance wasn’t loaded from any database. The using parameter will override the default.
Fields can be used to specify which fields to reload. The fields should be an iterable of field attnames. If fields is None, then all non-deferred fields are reloaded.
When accessing deferred fields of an instance, the deferred loading of the field will call this method.
- replace(data, run=None, format=None)¶
Replace artifact content.
- Parameters:
data (lamindb.core.types.UPathStr |
DataFrame|AnnData|MuData) – A file path.run (
Run|None, default:None) – The run that created the artifact gets auto-linked ifln.track()was called.
- Return type:
None
Examples
Say we made a change to the content of an artifact, e.g., edited the image
paradisi05_laminopathic_nuclei.jpg.This is how we replace the old file in storage with the new file:
>>> artifact.replace("paradisi05_laminopathic_nuclei.jpg") >>> artifact.save()
Note that this neither changes the storage key nor the filename.
However, it will update the suffix if it changes.
- restore()¶
Restore from trash.
Example:
artifact.restore()
- Return type:
None
- save(upload=None, **kwargs)¶
Save to database & storage.
- Parameters:
upload (
bool|None, default:None) – Trigger upload to cloud storage in instances with hybrid storage mode.- Return type:
Example:
import lamindb as ln artifact = ln.Artifact("./myfile.csv", key="myfile.parquet").save()
- save_base(raw=False, force_insert=False, force_update=False, using=None, update_fields=None)¶
Handle the parts of saving which should be done only once per save, yet need to be done in raw saves, too. This includes some sanity checks and signal sending.
The ‘raw’ argument is telling save_base not to save any parent models and not to do any changes to the values before save. This is used by fixture loading.
- serializable_value(field_name)¶
Return the value of the field name for this instance. If the field is a foreign key, return the id value instead of the object. If there’s no Field object with this name on the model, return the model attribute’s value.
Used to serialize a field’s value (in the serializer, or form output, for example). Normally, you would just access the attribute directly and not use this method.
- unique_error_message(model_class, unique_check)¶
- validate_constraints(exclude=None)¶
- validate_unique(exclude=None)¶
Check unique constraints on the model and raise ValidationError if any failed.
- view_lineage(with_children=True, return_graph=False)¶
Graph of data flow.
- Return type:
Digraph|None
Notes
For more info, see use cases: Data lineage.
Examples
>>> collection.view_lineage() >>> artifact.view_lineage()